Ontology 1o1¶
Introduction¶
In computer science, an ontology is a way of organizing knowledge in a structured and hierarchical manner. It consists of a set of classes or concepts, each with its own properties and relationships to other classes. Ontologies are used to help machines reason about information in a more intelligent and automated way, leading to more effective decision-making and problem-solving.
-- ChatGPT
Ontologies, as we know them in computer science, hail from the concept of ontology in philosophy, more precisely, the field of metaphysics.
There, "ontology" describes the doctrine of being and existence. It is mainly about the categorization of entities (which can be abstract or material), of processes and properties, and of events.
In this point, the philosophical concept of ontology agrees with that of computer science. The ontology of computer science, however, does not focus on philosophical issues but on representations of the categorized concepts and their relations to each other.
Ontologies have become more and more important in computer science since they allow for a standardized set of terms (so-called controlled vocabularies) which describe the things you are engaging with and putting them in a structure, guided by rules. This way, it becomes easier for humans to learn and communicate about a topic since ontologies also have a teaching nature on the one hand, and for machines to order these terms in a similar way to human understanding on the other hand.
Especially in the field of research data management, ontologies prove very useful because they achieve a compromise between human- and machine-readability and thus may lead to fewer miscommunications and errors. In the FAIR data principles, the I (for interoparability) can be achieved with the help of ontologies. This is done in, for example, the DataPLANT consortium.
Classes (& Instances)¶
Very similarly to OOP, classes are concepts (conceptual blueprints) of objects that can be instantiated (which results in an instance). If, e.g., there is the class "Cat", "Garfield" would be an instance of it.
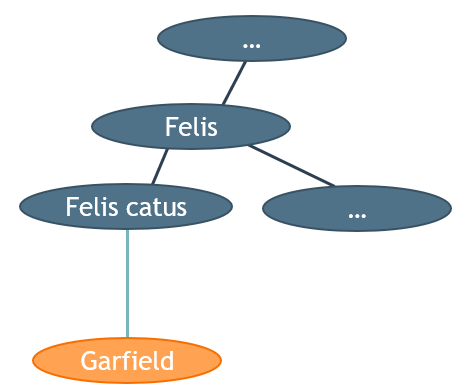
Classes can also have (an infinite amount of) subclasses. In the following graph, the domain of the kingdoms of life is (partially) represented as a class with subclasses:

An instance of the subclass "Felis catus" (Cat):
Relations¶
Objects in an ontology can be related to one another.
Beneath a set of ontology-specific individual relations, there are some relations that are seen in many ontologies or even every one:
- is_a: Describes the relation between a subclass and a class which leads to a child-term parent-term relation. The

- part_of: Describes the relation between subclasses and a class where the class can only be instantiated if every subclass is present (as instance). E.g. a "cell" cannot exist without all of its crucial parts, therefore "cell membrane" is part_of "cell".

- xref: Describes cross-reference between different ontologies. Often used for synonyms when dealing with multiple ontologies at once. E.g. "Organism" in OBI ontology can be xref of "organism" in NCBIT ontology.
Attributes¶
Attributes are properties that are related (via relations) to objects (classes and/or their instances). "Garfield" from above could have the attribute "orange fur" via a "has_a" relation.

Ontology file formats¶
There are many different file formats for ontologies. I will cover 2 of the most important (and most widely used) ones.
OWL¶
Web Ontology Language (OWL) is a knowledge representation language for ontologies. Its own file format has the extension .owl.
OWL uses different language implementations for its structure. Though there are several different ones, the most important ones are the XML and RDF implementations.
Here is an example of an ontology only consisting of the class "Felis_catus", its instance "Garfield" which has the attribute "orange_fur" via a "has_a" relation:
<?xml version="1.0"?>
<Ontology xmlns="http://www.w3.org/2002/07/owl#"
xml:base="http://www.semanticweb.org/revil/ontologies/2023/1/untitled-ontology-2"
xmlns:rdf="http://www.w3.org/1999/02/22-rdf-syntax-ns#"
xmlns:xml="http://www.w3.org/XML/1998/namespace"
xmlns:xsd="http://www.w3.org/2001/XMLSchema#"
xmlns:rdfs="http://www.w3.org/2000/01/rdf-schema#"
ontologyIRI="http://www.semanticweb.org/revil/ontologies/2023/1/untitled-ontology-2">
<Prefix name="" IRI="http://www.semanticweb.org/revil/ontologies/2023/1/untitled-ontology-2/"/>
<Prefix name="owl" IRI="http://www.w3.org/2002/07/owl#"/>
<Prefix name="rdf" IRI="http://www.w3.org/1999/02/22-rdf-syntax-ns#"/>
<Prefix name="xml" IRI="http://www.w3.org/XML/1998/namespace"/>
<Prefix name="xsd" IRI="http://www.w3.org/2001/XMLSchema#"/>
<Prefix name="rdfs" IRI="http://www.w3.org/2000/01/rdf-schema#"/>
<Declaration>
<Class IRI="#Felis_catus"/>
</Declaration>
<Declaration>
<ObjectProperty IRI="#has_a"/>
</Declaration>
<Declaration>
<NamedIndividual IRI="#Garfield"/>
</Declaration>
<Declaration>
<NamedIndividual IRI="#orange_fur"/>
</Declaration>
<ClassAssertion>
<Class IRI="#Felis_catus"/>
<NamedIndividual IRI="#Garfield"/>
</ClassAssertion>
<ObjectPropertyAssertion>
<ObjectProperty IRI="#has_a"/>
<NamedIndividual IRI="#Garfield"/>
<NamedIndividual IRI="#orange_fur"/>
</ObjectPropertyAssertion>
</Ontology>
While the OWL format is very mighty and due to XML easily parsable, it is not that simple to read and understand (as a human). Due to this, based on OWL, OBO has been founded.
OBO¶
OBO is a format developed by the Open Biological and Biomedical Ontologies Foundry (OBOFoundry). While this file format is based on the principles of OWL, it uses a vastly different language implementation in a slightly YAML style syntax. The same minimalistic ontology as in the OWL example, this time in OBO (flat file) format:
[Term]
id: GARFO:0000000
name: Felis_catus
is_a: OWL:0000000 ! Thing
[Instance]
id: GARFO:0000001
name: Garfield
instance_of: Felis_catus
property_value: has_a orange_fur
[Typedef]
id: GARFO:0000002
name: has_a
[Term]
id: GARFO:0000003
name: orange_fur
OBO is used as the primary format for ontologies in the DataPLANT consortium. OBO files have the extension .obo.
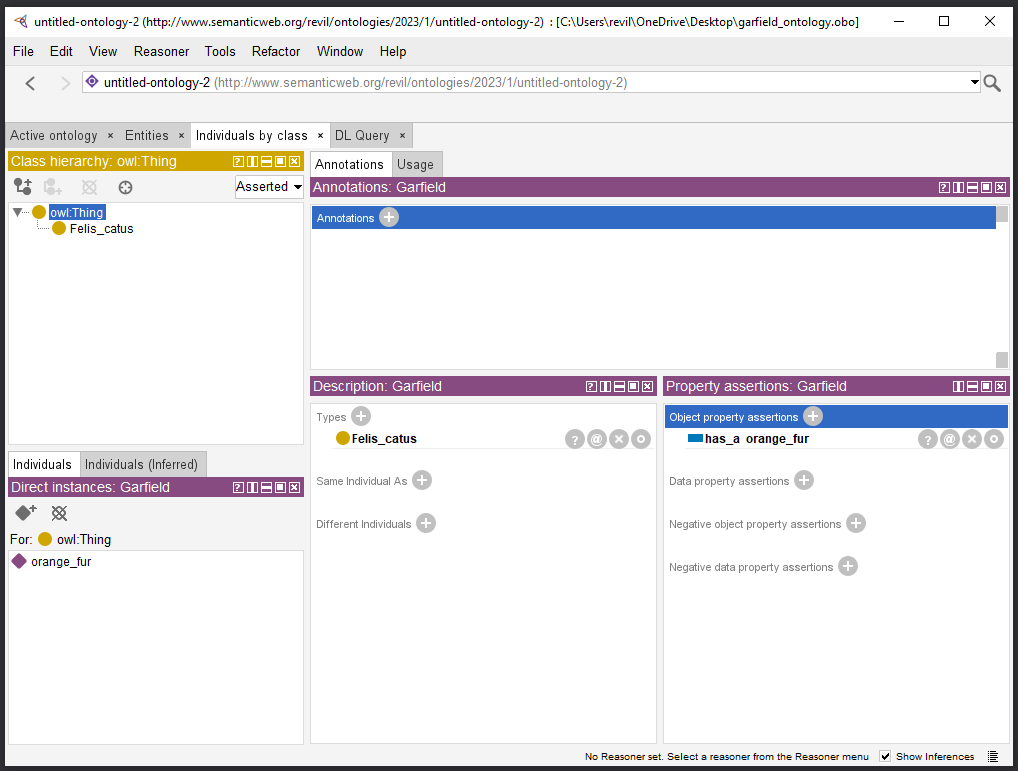
Protégé¶
Protégé is a free tool for creating ontologies. It supports reading OWL ontologies, creating and editing ontologies as well as exporting them in different formats (including OWL and OBO, although the latter one not in its easy-to-read flat file format).
Parsing OBO files in F¶
The BioFSharp library offers OBO parsing functionality:
#r "nuget: BioFSharp"
#r "nuget: BioFSharp.IO"
#r "nuget: FSharpAux"
open BioFSharp.IO
open FSharpAux.IO
let myTerm = Obo.createOboTerm "ID:0001" "myTerm" "" "" "" [] [] [] "" "" "" "" "" "" "" "" [] [] [] [] [] ""
myterm
- BioFSharp, 1.2.1
- BioFSharp.IO, 1.2.1
- fsharpaux, 1.0.0
| Id | Name | Namespace | Definition | Relationship | Related_synonym | IsA | Synonym | ExactSynonym | BroadSynonym | NarrowSynonym | XrefAnalog | Comment | IsObsolete | Replacedby | Consider | AltId | DisjointFrom | Subset | IntersectionOf | .. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
ID:0001 | myTerm | [ ] | [ ] | [ ] | [ ] | [ ] | [ ] | [ ] |
let fp = @"C:\Users\revil\OneDrive\Desktop\garfield_ontology.obo"
Seq.fromFile fp
|> Obo.parseOboTerms
| index | Id | Name | Namespace | Definition | Relationship | Related_synonym | IsA | Synonym | ExactSynonym | BroadSynonym | NarrowSynonym | XrefAnalog | Comment | IsObsolete | Replacedby | Consider | AltId | DisjointFrom | Subset | IntersectionOf | .. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | GARFO:0000000 | Felis_catus | [ OWL:0000000 ] | ||||||||||||||||||
| 1 | GARFO:0000003 | orange_fur |
Creating an ontology¶
First, you should model your ontology and ask yourself the questions "What do I want to cover?", "What is the scope of my ontology?" and begin with classes and subclasses, followed by relations and attributes. Instances should be covered at last.
Protégé helps you greatly with structuring and also visualizing it.
For implementation, I advice you to create the file of your ontology using Protégé or writing it manually (since OBO files are easy to write in their flat file format). A nice example can be seen in the DataPLANT broker ontology that gets updated regularly: https://github.com/nfdi4plants/nfdi4plants_ontology/blob/main/dpbo.obo
References¶
https://chat.openai.com/chat
https://en.wikipedia.org/wiki/Ontology_components
https://owlcollab.github.io/oboformat/doc/GO.format.obo-1_4.html
https://csbiology.github.io/BioFSharp//Obo.html