NB01a Systems Biology
This notebook introduces the field of Systems Biology and explains why programming is a necessary skill to it.
- Systems Biology: A brief introduction
- References
Systems Biology: A brief introduction
The term “systems theory” was introduced by the biologist L. von Bertalanffy. He defined a system as a set of related components that work together in a particular environment to perform whatever functions are required to achieve the system's objective (Bertalanffy 1945). The hierarchical organization orchestrating the interaction of thousands of molecules with individual properties allows complex biological functions. Biological processes like cell division, biomass production, or a systemic response to perturbations are molecular physiological functions which result from a complex dynamic interplay between genes, proteins and metabolites (Figure 1). To gain a holistic understanding of a biological system, all parts of the system need to be studied simultaneously by quantitative measures (Sauer et al. 2007). The focus on a system-wide perspective lies on the quantitative understanding of the organizational structure, functional state, robustness and dynamics of a biological system and led to the coining of the term “Systems Biology” (Kitano 2002a).
The current challenges of Systems Biology approaches are mainly along four lines (Sauer et al. 2007, Joyce and Palsson 2006):
(i) - system-wide quantification of transcriptome, proteome (including protein modifications) and metabolome
(ii) - identification of physical interactions between these components
(iii) - inference of structure, type and quantity of found interactions
(iv) - analysis and integration of the resulting large amounts of heterogeneous data. It becomes obvious that an interdisciplinary effort is needed to resolve these challenges in Systems Biology (Aderem 2006). Here Biology dictates which analytical, experimental and computational methods are required.
Modern analytical methods to measure the identity and quantity of biomolecules system-wide, summarized under the term “quantitative omics”-technologies, address the first two mentioned challenges of Systems Biology. Among these “omics”-technologies are transcriptomics based on microarrays/next generation sequencing and proteomics/metabolomics based on mass-spectrometry.
Tying in with the area of genome sequencing, the focus is set on the accurate profiling of gene/protein expression and metabolite concentrations, as well as on the determination of biological protein modifications and of physical interactions between proteins.
Addressing the abovementioned challenges three and four of Systems Biology, the development of numerous computational approaches reaches out to unravel the intrinsic complexity of biological systems (Kahlem and Birney 2006). These computational approaches focus on knowledge discovery and on in silico simulation or modeling (Kitano 2002b). In the latter approach knowledge on a biological process is converted into a mathematical model. In silico simulations based on such a model can provide predictions that may subsequently be tested experimentally. Computation-based knowledge discovery (also known as data mining) aims to extract hidden patterns from complex and high-dimensional data to generate hypotheses. Therefore, the first step is to describe information on a biological system such that it is sustainably stored in a format rendering it readable and manipulable for machines and humans. The second step is to integrate the huge amount of differently structured data, often referred to as the “big data” challenge. In a last step, statistical or machine learning methods are applied to extract the information or underlying principles hidden in the data.
The most flexible way of working with huge amounts of data is using a lightweight programming language with a succinct syntax. Therefore, it becomes necessary that biologist become familiar with a suitable programming language to solve real world problems in (Systems) Biology.
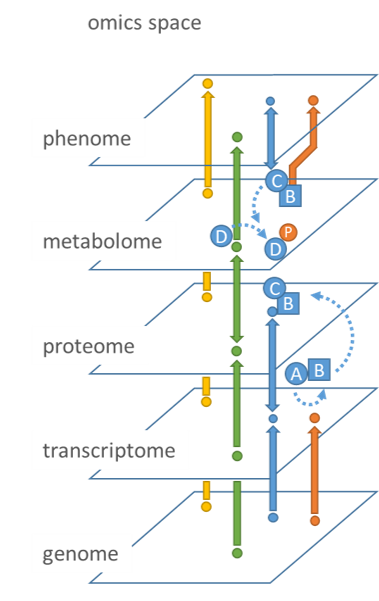
Figure 1: A conceptual view of the omic space.
The omics space comprises of genomic, transcriptomic, proteomic, metabolomic and phenomic systems level represented as a plane. Complex biological function is the result of the interplay between molecules of one and/or different systems level.
References
- Bertalanffy, L. von. Zu einer allgemeinen Systemlehre. Blätter für deutsche Philosophie 18 (1945).
- Sauer, U., Heinemann, M. & Zamboni, N. Genetics. Getting closer to the whole picture. Science 316, 550–551; 10.1126/science.1142502 (2007).
- Kitano, H. Systems biology. a brief overview. Science 295, 1662–1664; 10.1126/science.1069492 (2002).
- Joyce, A. R. & Palsson, B. O. The model organism as a system. integrating 'omics' data sets. Nat Rev Mol Cell Bio 7, 198–210; 10.1038/Nrm1857 (2006).
- Aderem, A. Systems biology. Its practice and challenges. Cell 121, 511–513; 10.1016/j.cell.2005.04.020 (2005).
- Kahlem, P. & Birney, E. Dry work in a wet world. computation in systems biology. Mol Syst Biol 2 (2006).
- Kitano, H. Computational systems biology. Nature 420, 206–210; 10.1038/nature01254 (2002).