The ProteomIQon
The ProteomIQon is a collection of open source computational proteomics tools to build pipelines for the evaluation of MS derived proteomics data written in F#. The current state of the tool chain allows handle tasks like signal detection, peptide identification, quantification and protein inference.
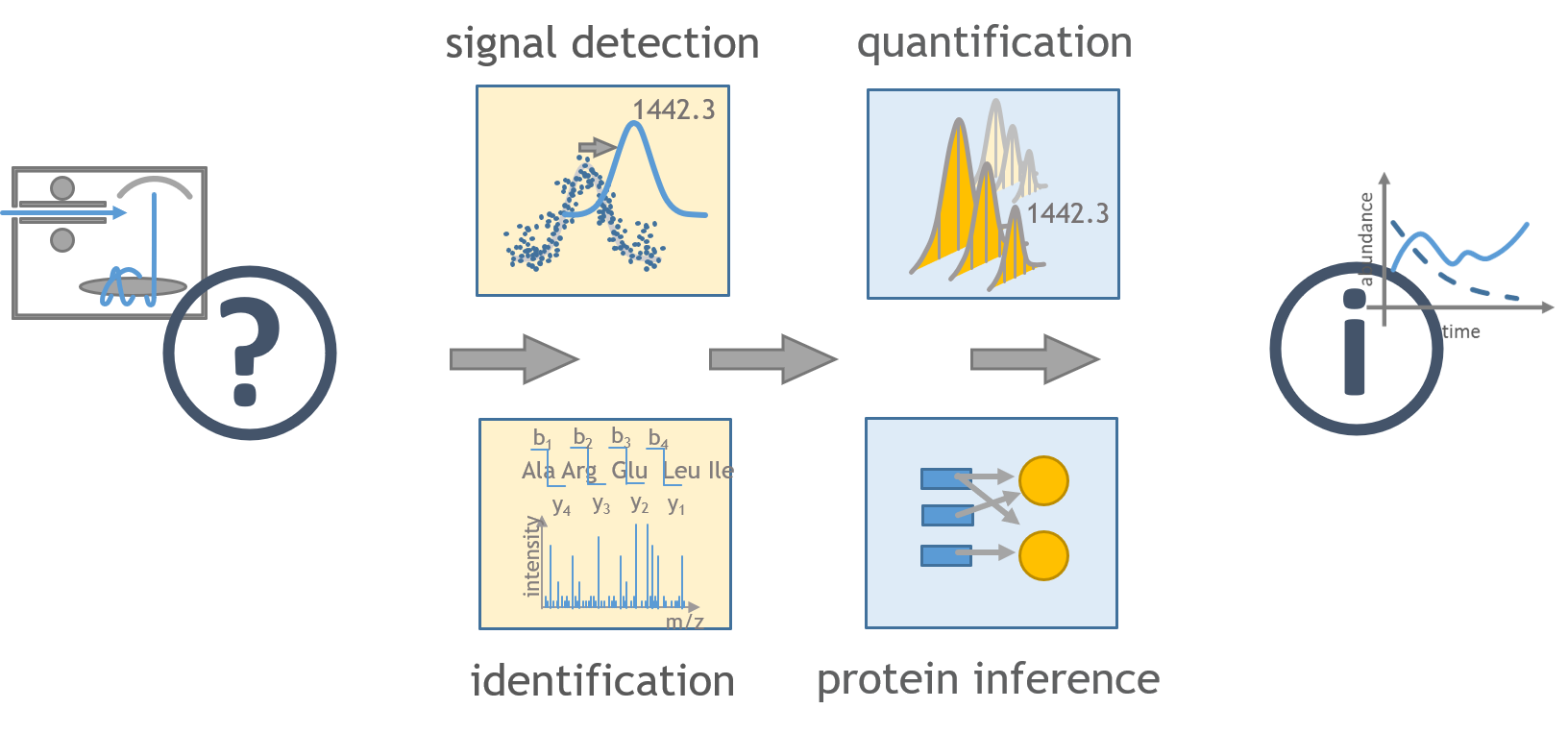
Each ProteomIQon tool is concerned with a specific task. This makes the tool-chain flexibel and easily extendable. An example of a prototypical chaining of tools to identify and quantify a mix of 14N and 15N labeled proteins can be found in the here. We are currently working on the cwl tool and workflow descriptions, so you can expect a to see more workflow graphs in the near future!
All tools are available using nuget and soon via BioConda. Each tool is described in detail on its corresponding documentation page, accessible via the navigation pane, if you think a functionality is missing, feel free to contact us or to join us as a contributor!
The Core Project
The ProteomIQon core is referenced by all tools. It contains mainly serializable data transfer objects such as tool results and tool parameters, as well as their mapping to domain specific types. This is also the place for any kind of code reusable across tools such as thin wrappers around data readers, logging or CLI formatting.
Documentation
The documentation and tutorials for this library are automatically generated (using the F# Formatting) from .fsx and .md files in the docs folder. If you find a typo, please submit a pull request!
Contributing
Please refer to the CSB Contribution guidelines
Community/Social
Want to get in touch with us? We recently joined the twitter crowd:
.svg)

